One-step versus two-step PCR? The 12,000 pound question
The multiplex PCR strategy we had decided to focus on for the Zibra trip was certainly going to be one of the most technically challenging parts of process. Not least because we are genomics group who grew up in the era of NGS and have very little experience tweaking PCR. Others have been reporting difficulty sequencing Zika genomes. This is likely bbecause of the low viral load found in clinical samples. This results in low numbers of reads using a direct metagenomics approach, meaning a huge amount of sequencing is required to get satisfactory genome coverage. When we started sequencing Ebola genomes using MinION in Guinea we favoured amplicon sequencing because it made uploading the data via a 3G connection slightly less painful, as PCR enriches for “on-target” reads. Back then we used a Superscript III one-step RT-PCR kit which anyone will tell you is brilliantly simple and Simon Weller at DSTL had already shown proof of principle data that we could do two or three reactions in multiplex using it on Ebola.
In one-step RT-PCR the idea is to do an initial reverse transcription step off the reverse primer and then proceed to PCR cycling. You therefore need an enzyme cocktail of reverse transcriptase and a DNA polymerase along with a reaction buffer optimised for both reactions. The benefit of this approach is obviously simplicity and people do multiplex qRT-PCR routinely so it seemed like a good option to explore. The alternative is a two-step approach where you have a separate cDNA synthesis step, which typically uses poly-T, random hexamers or specific primers for reverse transcription. This approach would give us the flexibility to use different priming strategies for each step i.e. random primers for the RT and multiplex specific primers for the PCR. Typically you don’t clean up the first step reaction but keep the input to the second step to below 10% volume.
We had to make a decision on what reagents to order from NEB as time was rapidly slipping by until the start of the trip. I decided to design a quick bake-off to determine which was better suited to multiplexing and ultimately our Zika protocol. Using two-step reagents (as you can’t really do it the other way round) this experiment would compare random hexamers with specific primers for reverse transcription with a toy multiplex PCR of five primer pairs at three concentrations for the second step giving a total of six conditions which I barcoded with the native barcoding kit and sequenced on the MinION. If specific primers outperformed random primers we would order one-step reagents and if random hexamers triumphed we would order two-step kits. It was a decision that could leave us with £12k of the wrong kits. A straw pole had Nick backing random hexamers and me specific primers. Nick was right.
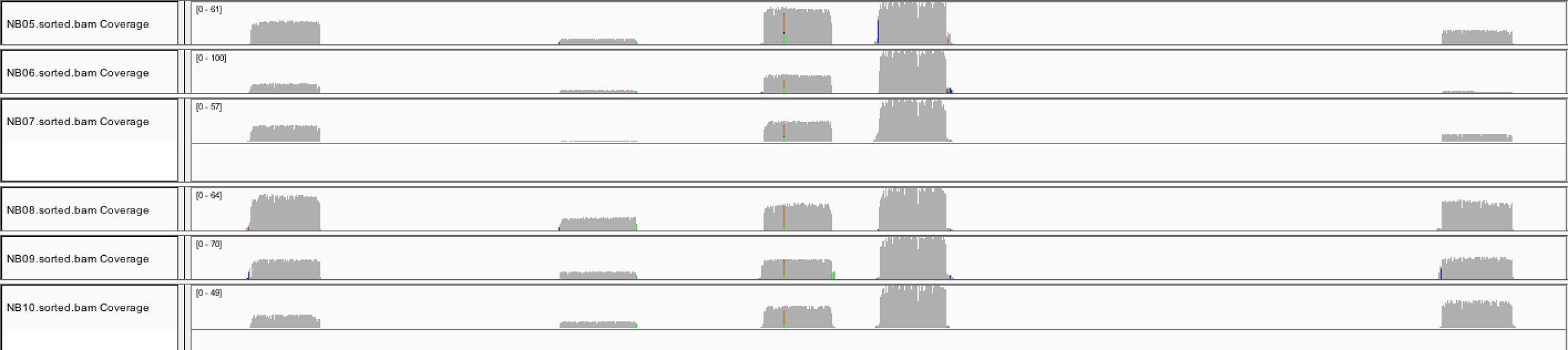
Experimental Conditions
| Name | Primers for cDNA | Primers for PCR |
|---|---|---|
| NB05 | Specific primers | 0.05 uM each primer |
| NB06 | Specific primers | 0.025 uM each primer |
| NB07 | Specific primers | 0.0125 uM each primer |
| NB08 | Random hexamers | 0.05 uM each primer |
| NB09 | Random hexamers | 0.025 uM each primer |
| NB10 | Random hexamers | 0.0125 uM each primer |
It appeared that random priming during the reverse transcription step gave a more even representation of the five products in the second step. Region two for example is better represented at lower primer concentrations when using random primers for the first step, as is region two to a lesser extent. It is possible this is is caused by uneven cDNA synthesis due to interactions between the multiplex primers at 50°C, the optimum temperature for reverse transcriptase.
In a follow-up experiment I wanted to see if I could balance the representation by increasing the primer concentration for the weakest product (2) and reducing the concentration for the strongest (5). As you can see below that works quite nicely with all products present at fairly equal abundances. This would a useful technique for tuning the abundances in the final Zika scheme although that would be a late optimisation.

R9_ZKV Random hexamers 0.05 uM regions 1+3+5, 0.1 uM region 2 and 0.025 uM region 4
Overall I was quite happy with our first tests doing multiplex PCR, I felt what we had was enough to justify ordering the kits so we quickly sent an order over to Chris Lounds at NEB with about a week to go before the trip. We heard back that the reagents would arrived the next day apart from the Blunt/TA ligase which would have to come from the US as we had exceeded UK stock levels with our order of 15 packs.
