About this project
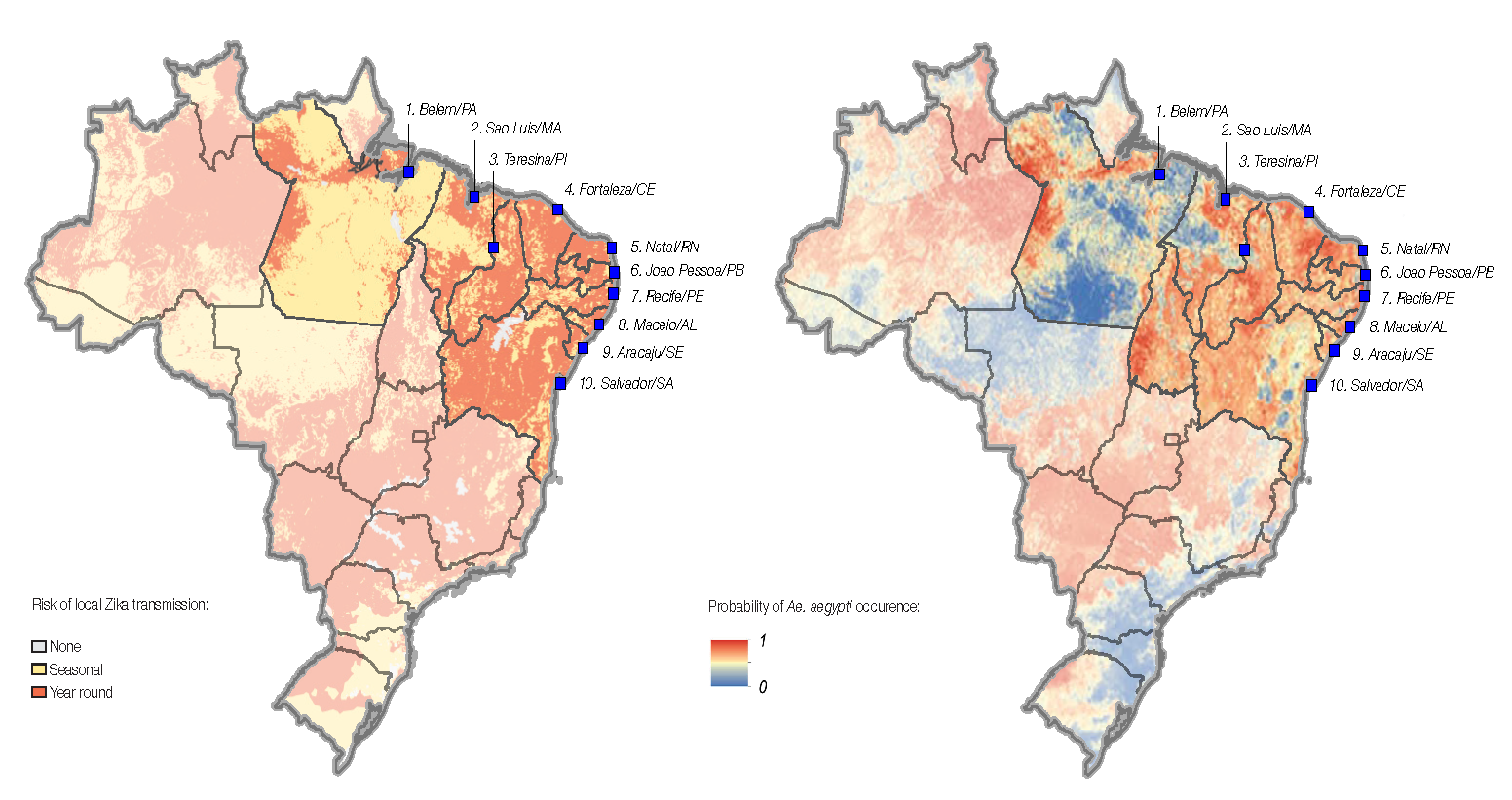
A major epidemic of Zika virus (ZIKV) is ongoing in Latin America. Critically, there is evidence for an association between infection with ZIKV and both microcephaly in newborns and Guillain-Barré syndrome. Currently there is a paucity of complete genome sequences for this virus, in part due to difficulties in transporting material outside of Brazil for sequencing, hindering attempts to determine virus origins, epidemiology and any genomic basis to microcephaly.
We have recently established real-time portable genome sequencing using the Oxford Nanopore MinION device, and successfully used this to characterize Ebola virus genetic diversity in Guinea during the 2014-2015 outbreak.
We propose to extend this ground-breaking achievement to ZIKV by establishing two portable genome sequencing laboratories in Brazil. Through collaboration with the Oswaldo Cruz Foundation (FIOCRUZ) and the Instituto Evandro Chagas public health laboratory in Brazil we will sequence 750 complete genomes of ZIKV, covering a broad geographical region including historical samples, and from patients with a range of clinical presentations. These novel genomic data will provide key information on how and when ZIKV was introduced to Brazil, the pattern and determinants of spread through the country and to neighbouring localities, the extent of genetic diversity (of importance to vaccine and diagnostic design), and whether there are any associations between changes in the virus genome and the likelihood of ZIKV complications such as microcephaly.
Crucially these data will provide a surveillance framework for tracking further spread into other geographic regions. In common with our previous efforts, this effort will serve as a beacon for open science during a public health emergency.
Data will be subject to open release as it is generated.